Ruben Wolff

x.com/Ruben_Wolff
bsky.app/profile/rubenwolff.bsky.social
linkedin.com/in/ruben-wolff
mastodon.social/@ruben_wolff
anchor.fm/ruben-wolff
bsky.app/profile/rubenwolff.bsky.social
linkedin.com/in/ruben-wolff
mastodon.social/@ruben_wolff
anchor.fm/ruben-wolff
Independent Consulting
- Designed and lead the implementation team for a new enterprise wide real time message bus. The new system could auto-scale up to 30000 messages per second which could be read via streaming, big data bulk or API access with full access control over active directory. Real time Machine learning models where utilized to align multiple IOT sensors for most accurate ball and player tracking as well as player action classification.
- Consulting Client: Sports events company in the top 5 worldwide
- Prototyped a series of models to improve "eDiscovery" like work in the Swiss legal context for one of the countries largest white collar crime cases. Models included NLI (sentence contradiction / agreement classification), semantic document similarity search, document classification image+text and simple OCR.
- Client: Initially a Swiss law firm but now the project as spun out into a separate company competing with Relativity in the Swiss market
- Machine learning advisor for a oracle node reputation scoring system.
- Client: small community governed oracle network on Binance smartchain
- Advanced predictive risk models (for NEC, ROP, IVH, CLD, and Sepsis) for the neonatal ICU.
- Client: One of the largest Neonatal care providers in the USA
- Length of Stay prediction in burn patients.
- Client: Altonaer Children’s Hospital, Hamburg. (Related Publication)
Past Startups
- Corepo
 is a knowledge graph generation project which performs monthly whole web scans to identify all companies with a web presence. A machine learning system identifies which sites are company homepages followed by a deeper crawl and information extraction to identify metadata such as founding year, location, industry and relevant employees. The B2C end user product is designed to bridge the gap between the limitations of human curated company databases and general purpose web search engines.
is a knowledge graph generation project which performs monthly whole web scans to identify all companies with a web presence. A machine learning system identifies which sites are company homepages followed by a deeper crawl and information extraction to identify metadata such as founding year, location, industry and relevant employees. The B2C end user product is designed to bridge the gap between the limitations of human curated company databases and general purpose web search engines.
- Funding: Angel
- Conclusion: B2B data access agreements achieved revenue stage. Operations and sales has been handed off to a third party.
 the AI that reads pubmed for you. This NLP project aimed to create automated literature review on the fly every day as new publications appear on pubmed.gov and clinicaltrials.gov. The target customers were medical directors in big pharma who need to make daily decisions on a diversity of drugs and conditions and have little time to read but also can not rely on the always out of date published literature reviews. The machine learning model first performs named entity recognition for interventions(drugs), conditions(MeSH) and outcome measures (such as mOS, PFS etc.) followed by a linking model to group together the outcome number to the appropriate study arm. This first version of the system was focused on clinical oncology only.
the AI that reads pubmed for you. This NLP project aimed to create automated literature review on the fly every day as new publications appear on pubmed.gov and clinicaltrials.gov. The target customers were medical directors in big pharma who need to make daily decisions on a diversity of drugs and conditions and have little time to read but also can not rely on the always out of date published literature reviews. The machine learning model first performs named entity recognition for interventions(drugs), conditions(MeSH) and outcome measures (such as mOS, PFS etc.) followed by a linking model to group together the outcome number to the appropriate study arm. This first version of the system was focused on clinical oncology only.
- Funding: Entrepreneur First
- Conclusion: No product market fit; unlike our early adopters the broader market expected a level of accuracy and granularity only possible by full human curation
- Neoglia LTD
 was a London based health tech startup which I co-founded with the goal of increasing the utility of sensitive medical data siloed in hospital EHR. We prototyped a federated machine learning approach where clinical data would remain within the individual hospitals servers while allowing pharma companies to build models on the data from all participating hospitals.
was a London based health tech startup which I co-founded with the goal of increasing the utility of sensitive medical data siloed in hospital EHR. We prototyped a federated machine learning approach where clinical data would remain within the individual hospitals servers while allowing pharma companies to build models on the data from all participating hospitals.
- Funding: Entrepreneur First
- Conclusion: No product market fit; most major players had unpublicized alternative means of accessing sensitive data via research collaborations or curated anonymized downloadable datasets
Past Consulting with 
- Data Architecture Big Data for Stock Exchange
- Distributed Algorithm for Order Book state reconstruction from historical trade data
- Medical Claim Embedding Space from Medical Insurance Metadata
- Medical Records Diabetes Risk Prediction
- Medical Insurance Claims Adjudication Automation
- Claim Processing Time/Complexity prediction
- Automate Model refresh loop of Productive Machine learning Model (Data acquisition, Model training, Model Evaluation, Hot swap productive model behind live API)
- Data warehouse Modeling and Generation Package
- Data Architecture Data warehouse Belgium Health insurance
- Deployment Automation Data warehouse
Past Jobs
- Data Science intern at
investigating market behavior time series datasets and developing algorithmic trading strategies
- Software Developer at
 developing a Personalized Oncology Drug Suggestion Tool
developing a Personalized Oncology Drug Suggestion Tool - Neuroscience Researcher at
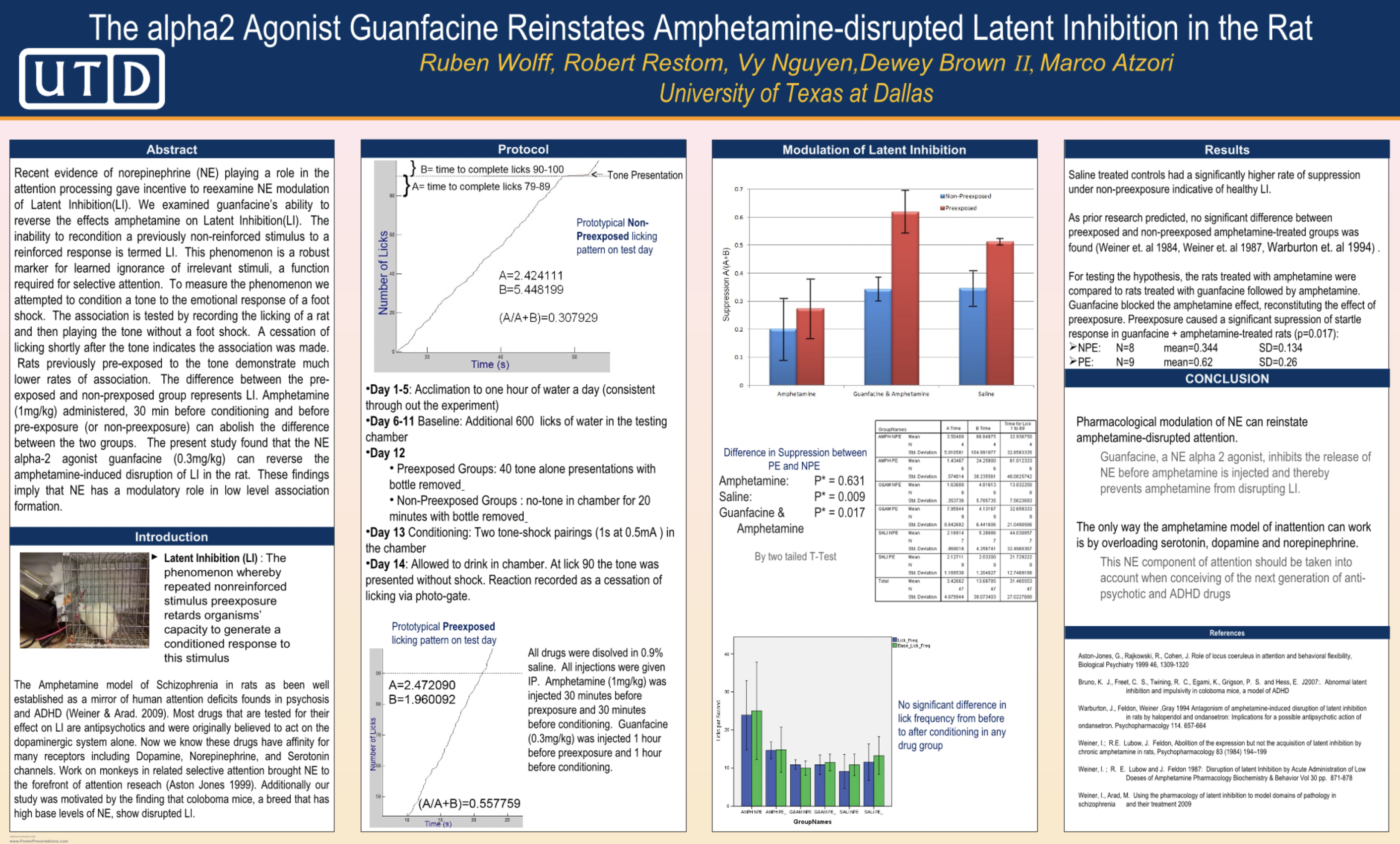
 leading a Guanfacine Selective Attention inVivo Study
leading a Guanfacine Selective Attention inVivo Study
Education
Master Thesis at  Under Thomas Hoffmann
Under Thomas Hoffmann
Distributed Structured Prediction For 3D Image Segmentation
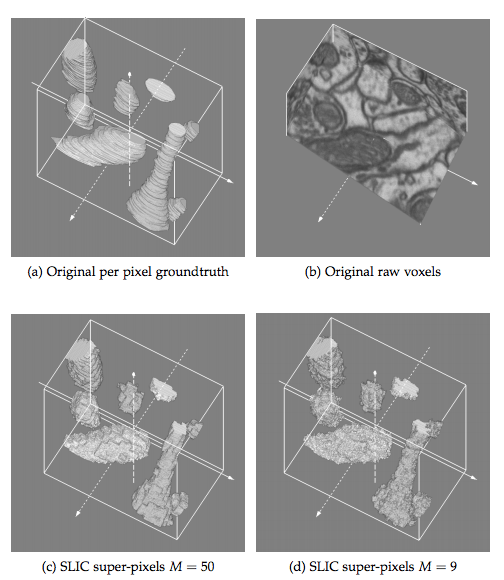
Bachelor Thesis at  Under Marco Atzori
Under Marco Atzori
Thesis: The α2 Agonist Guanfacine Reinstates Amphetamine-Disrupted Latent Inhibition in Rats
Related :
a-Adrenergic receptors in auditory cue detection: a2 receptor blockade suppresse false alarm responding in the rat
Past Talks
- Automating insurance claim processing: Using NLP methods to learn sparse spaces - at - SDS 2018 Slides
- The surprising Utility of NLP algorithms on non Text data - at - SwissText 2018 - agenda
- Machine Learning mustn't be a Black box - at - Certificate of Advanced Studies ZFH in Claims Management (12 ECTS) - slides
- Real Life Benchmarking - at - SQL server 2016 launch event - slides
- Digitize the Data Store - at - Common Sense 1702 - slides - video
Research
Peerbench.ai: Z. Cheng, R. Wolff et al., “Benchmarking Is Broken—Don’t Let AI Be Its Own Judge,” in Proc. 39th Conf. Neural Information Processing Systems (NeurIPS), 2026.
Preprint: https://arxiv.org/abs/2510.07575
NeurIPS Virtual Poster: https://neurips.cc/virtual/2025/loc/san-diego/poster/121919